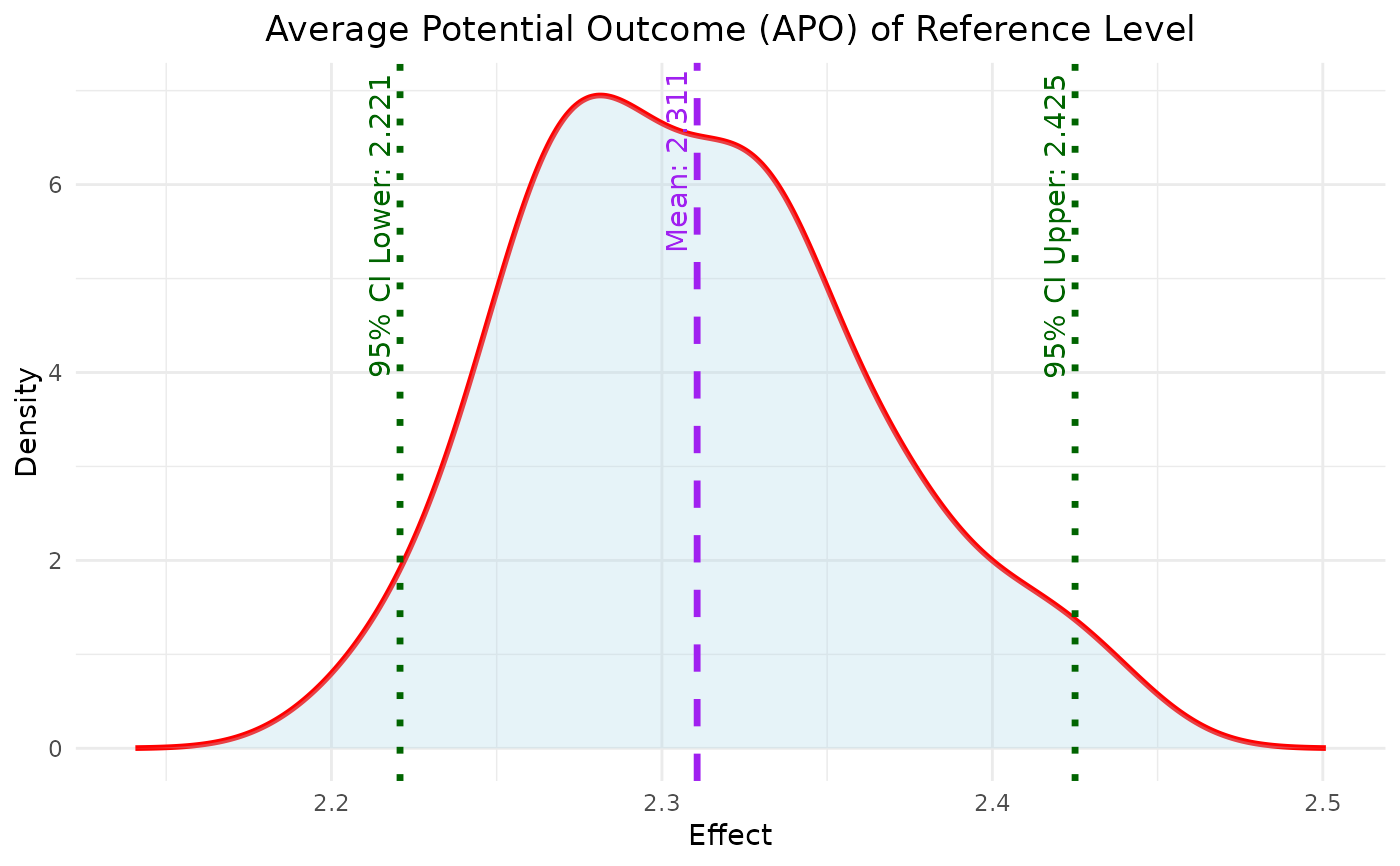
This function plots the density of APO for a specified effect type from bayesmsm output.
Value
A ggplot object representing density plot showing the distribution of the specified average potential outcome (reference or comparison).
Examples
# 1) Specify simple treatment‐assignment models
amodel <- list(
c("(Intercept)" = 0, "L1_1" = 0.5, "L2_1" = -0.5),
c("(Intercept)" = 0, "L1_2" = 0.5, "L2_2" = -0.5, "A_prev" = 0.3)
)
# 2) Specify a continuous‐outcome model
ymodel <- c("(Intercept)" = 0,
"A1" = 0.2,
"A2" = 0.3,
"L1_2" = 0.1,
"L2_2" = -0.1)
# 3) Simulate without right‐censoring
testdata <- simData(
n = 200,
n_visits = 2,
covariate_counts = c(2, 2),
amodel = amodel,
ymodel = ymodel,
y_type = "continuous",
right_censor = FALSE,
seed = 123)
model <- bayesmsm(ymodel = Y ~ A1 + A2,
nvisit = 2,
reference = c(rep(0,2)),
comparator = c(rep(1,2)),
treatment_effect_type = "sq",
family = "binomial",
data = testdata,
wmean = rep(1,200),
nboot = 10,
optim_method = "BFGS",
seed = 890123,
parallel = FALSE)
plot_APO(model$bootdata, effect_type = "effect_comparator")
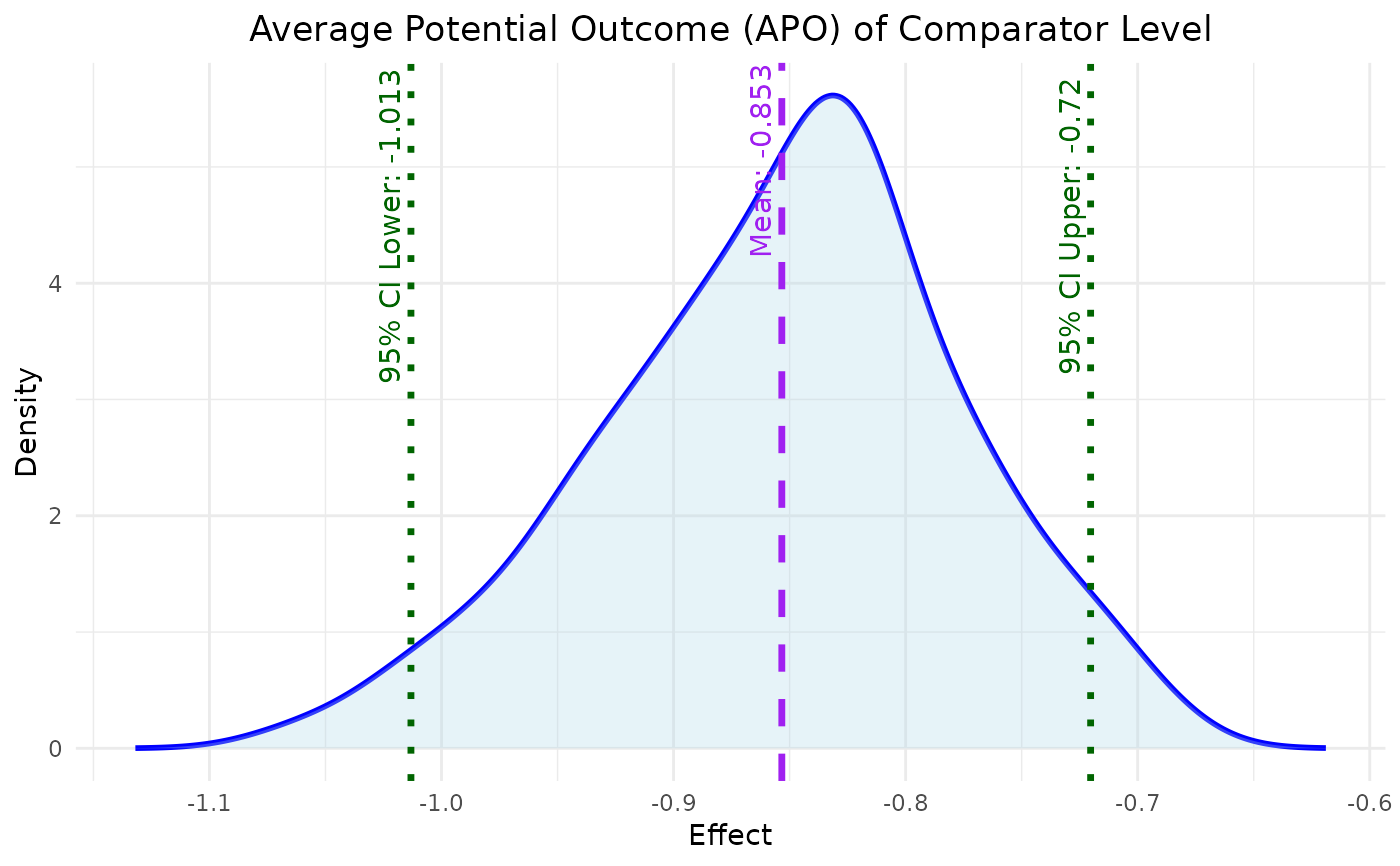 plot_APO(model, effect_type = "effect_reference")
plot_APO(model, effect_type = "effect_reference")