
causens: an R package for causal sensitivity analysis methods
Source:vignettes/causens-vignette.Rmd
causens-vignette.RmdIntroduction
Methods to estimate causal quantities often rely no unmeasured
confounding. However, in practice, this is often not the case.
causens is an R package that provides methods to estimate
causal quantities in the presence of unmeasured confounding. In
particular, causens implements methods from the three
following school of thoughts:
- Frequentist methods based on sensitivity functions
- Bayesianism via parametric modelling
- Monte Carlo sensitivity analysis
All implemented methods are accessible via the causens
function with the method parameter. The package also
provides a simulate_data function to generate synthetic
data on which to test the methods.
Installation
The package can be installed from GitHub via
devtools.
library(devtools)
devtools::install_github("Kuan-Liu-Lab/causens")Methods
Summary of the Unmeasured Confounder Problem
In causal inference, the potential outcome framework is often used to define causal quantities. For instance, if a treatment is binary, we respectively define and as the outcomes under treatment and control as if we can intervene on them a priori. However, in observational settings, this is often not the case, but we can still estimate estimands, e.g. the average treatment effect , using observational data using causal assumptions that relate potential outcome variables to their observational counterpart:
- Consistency:
- Conditional exchangeability:
- Positivity:
where is a set of observed confounders that can be used to adjust for confounding. In practice, it is often difficult to know whether these assumptions hold, and in particular, whether all confounding variables are contained in .
Hereon, we define
,
the set of unmeasured confounders, although, in causens, we
assume
to be univariate for simplicity.
Simulated Data Mechanism
We posit the following data generating process:
Using consistency, we define
.
Note that, in simData.R, we provide some options to allow
the simulation procedure to be more flexible. Parameters
and
dictate how the unmeasured confounder
affects the treatment
and the outcome
,
respectively; by default, they are set to 0.2 and 0.5.
Frequentist Methods (Brumback et al. 2004, Li et al. 2011)
Building on the potential outcome framework, the primary assumption that requires adjustment is conditional exchangeability. First, the latent conditional exchangeability can be articulated as follows to include :
Secondly, we can define the sensitivity function , with being a valid treatment indicator and being a propensity score value.
When there are no unmeasured confounders, for all and since .
Given a valid sensitivity function, we can estimate the average treatment effect via a weighting approach akin to inverse probability weighting.
For parsimony, we provide an API for constant and linear sensitivity functions, but we eventually will allow any valid user-defined sensitivity function.
# Simulate data
data <- simulate_data(
N = 10000, seed = 123, alpha_uz = 1,
beta_uy = 1, treatment_effects = 1
)
# Treatment model is incorrect since U is "missing"
causens_sf(Z ~ X.1 + X.2 + X.3, "Y", data = data,
c1 = 0.25, c0 = 0.25)$estimated_ate
#> [1] 1.005025
plot_causens(Z ~ X.1 + X.2 + X.3, data, "Y",
c1_upper = 0.5, c1_lower = 0, r = 1, by = 0.01)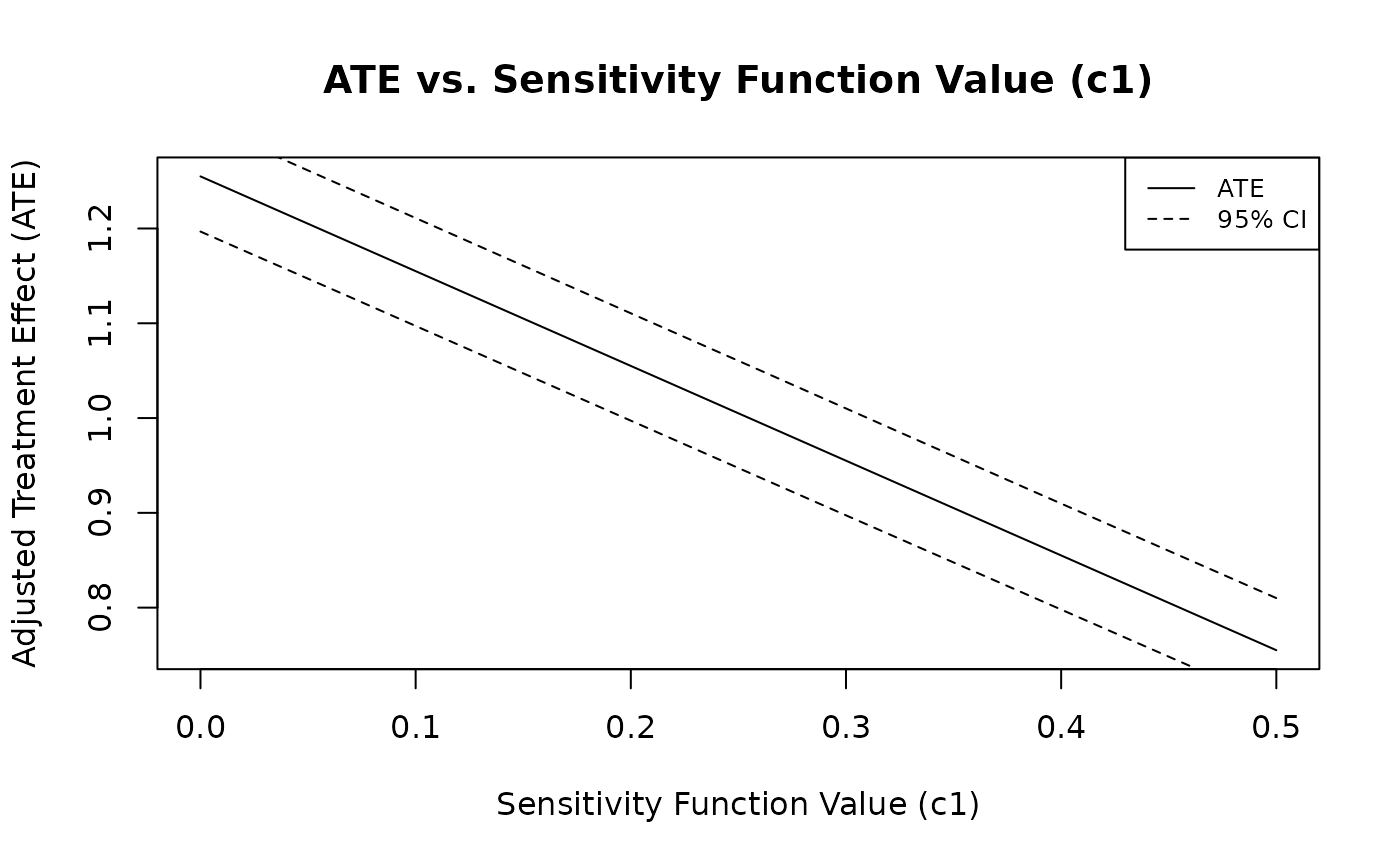
Bayesian Methods
In Bayesianism, we can adjust for unmeasured confounding by explicitly modelling and its relationship with and . Using a JAGS backend to carry out the MCMC procedure, we can estimate the average treatment effect by then marginalizing over . We assume the following data-generating mechanism and Bayesian model in causens:
data <- simulate_data(
N = 1000, alpha_uz = 0.5, beta_uy = 0.2,
seed = 123, treatment_effects = 1,
y_type = "continuous"
)
bayesian_causens(
Z ~ X.1 + X.2 + X.3, Y ~ X.1 + X.2 + X.3,
U ~ X.1 + X.2 + X.3, data
)Monte Carlo Approach to Causal Sensitivity Analysis
Warning Only works for binary outcomes, for now.
The Monte Carlo approach proceeds through the following steps:
- Produce a “naive” model where is ignored: .
- With estimates and of and obtained from fitting the naive model, we can sample from an assumed prior distribution:
- For each sample indexed by , we can compute bias adjusted estimates:
- Finally, we can sample the bias adjusted average treatment effect estimate to account for errors incurred in previous steps. With , we can compute the Monte Carlo estimate of the average treatment effect by taking their average.
data <- simulate_data(
N = 1000, alpha_uz = 0.2, beta_uy = 0.5,
seed = 123, treatment_effects = 1, y_type = "binary",
informative_u = FALSE
)
causens_monte_carlo("Y", "Z", c("X.1", "X.2", "X.3"), data)$estimated_ate
#> [1] 0.9929688